
Anisotropic models
David Bolin and Alexandre B. Simas
Created: 2024-10-23. Last modified: 2026-01-29.
Source:vignettes/anisotropic.Rmd
anisotropic.RmdIntroduction
For domains
,
the rSPDE package implements the anisotropic Matérn model
Where
is a
positive definite matrix,
and
is a constant chosen such that
would have the covariance function
if the domain was
,
i.e., a stationary and anisotropic Matérn covariance function. The
matrix
is defined as
with
and
.
Implementation details
Let us begin by loading some packages needed for making the plots
We start by creating a region of interest and a spatial mesh using
the fmesher package:
library(fmesher)
n_loc <- 2000
loc_2d_mesh <- matrix(runif(n_loc * 2), n_loc, 2)
mesh_2d <- fm_mesh_2d(
loc = loc_2d_mesh,
cutoff = 0.01,
max.edge = c(0.1, 0.5)
)
plot(mesh_2d, main = "")
We now specify the model using the matern2d.operators
function.
sigma <- 1
nu <- 0.5
hx <- 0.08
hy <- 0.08
hxy <- 0.5
op <- matern2d.operators(
hx = hx, hy = hy, hxy = hxy, nu = nu,
sigma = sigma, mesh = mesh_2d
)The matern2d.operators object has
cov_function_mesh method which can be used evaluate the
covariance function on the mesh. For example
r <- cov_function_mesh(op, p = matrix(c(0.5, 0.5), 1, 2))
proj <- fm_evaluator(mesh_2d, dims = c(100, 100), xlim = c(0, 1), ylim = c(0, 1))
r.mesh <- fm_evaluate(proj, field = as.vector(r))
cov.df <- data.frame(
x1 = proj$lattice$loc[, 1],
x2 = proj$lattice$loc[, 2],
cov = c(r.mesh)
)
ggplot(cov.df, aes(x = x1, y = x2, fill = cov)) +
geom_raster() +
xlim(0, 1) +
ylim(0, 1) +
scale_fill_viridis()
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).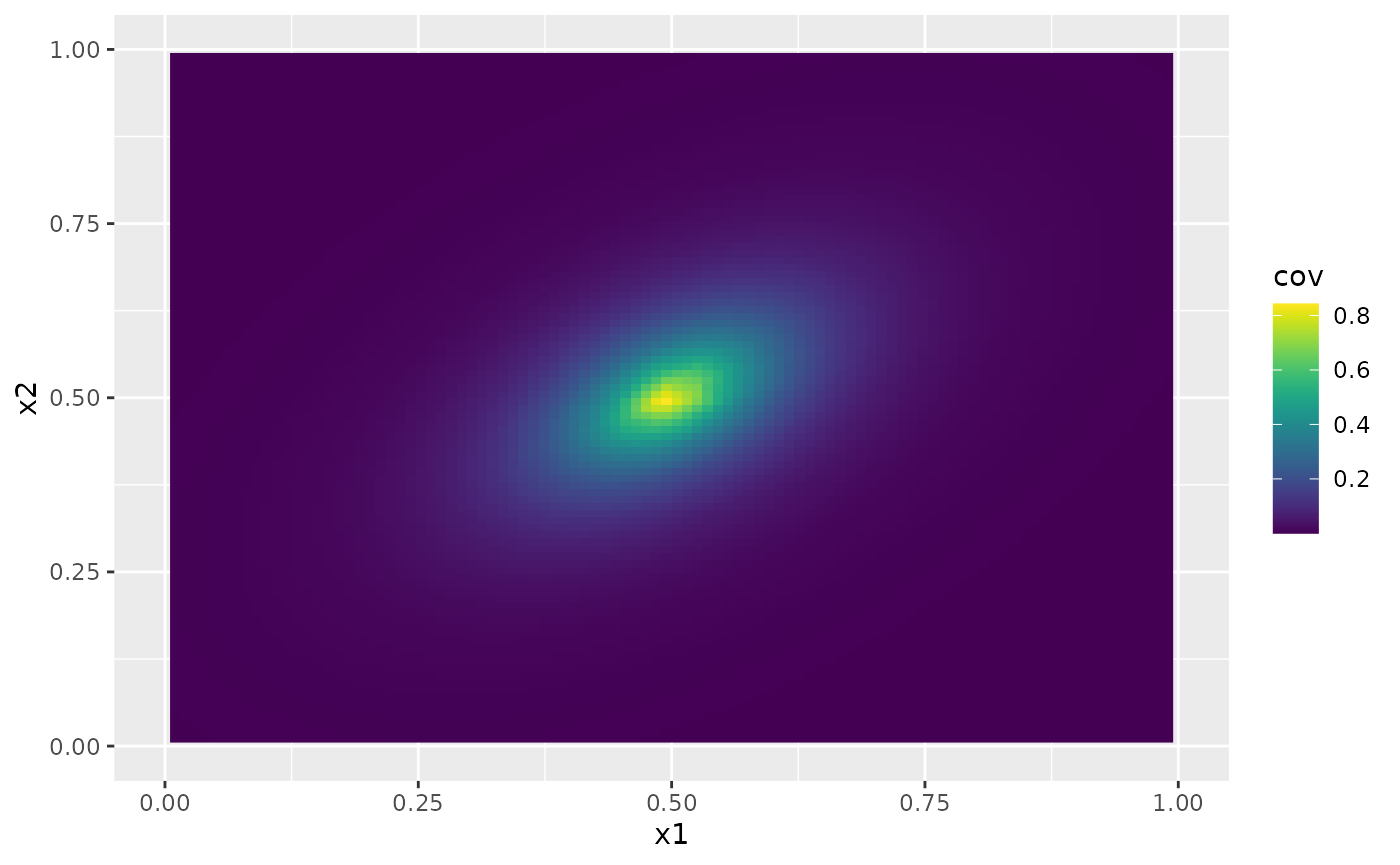
We can simulate from the field using the
simulate method:
u <- simulate(op)
proj <- fm_evaluator(mesh_2d, dims = c(100, 100), xlim = c(0, 1), ylim = c(0, 1))
u.mesh <- fm_evaluate(proj, field = as.vector(u))
cov.df <- data.frame(
x1 = proj$lattice$loc[, 1],
x2 = proj$lattice$loc[, 2],
u = c(u.mesh)
)
ggplot(cov.df, aes(x = x1, y = x2, fill = u)) +
geom_raster() +
xlim(0, 1) +
ylim(0, 1) +
scale_fill_viridis()
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).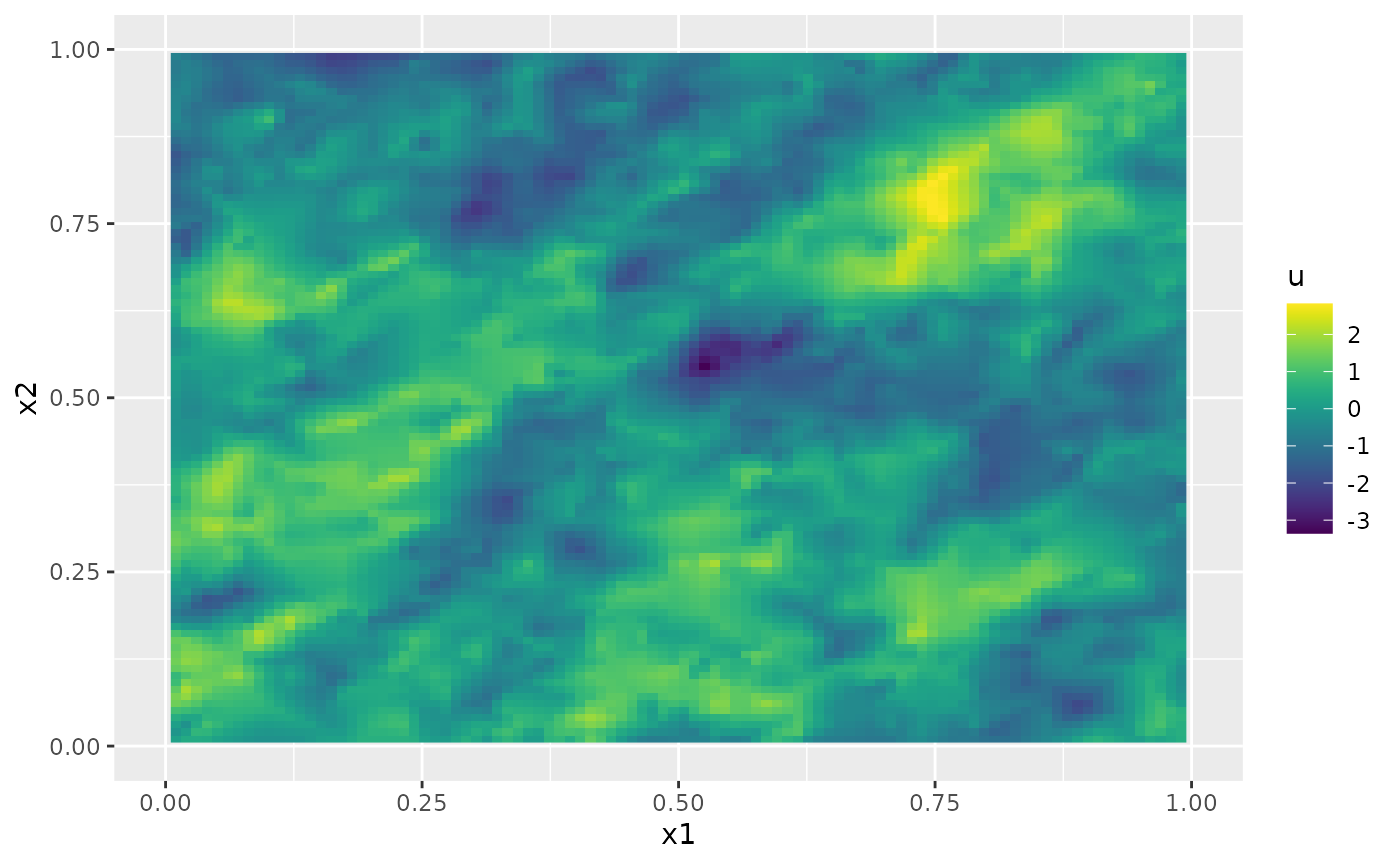
Let us now simulate some data based on this simulated field.
n.obs <- 2000
obs.loc <- cbind(runif(n.obs), runif(n.obs))
A <- fm_basis(mesh_2d, obs.loc)
sigma.e <- 0.1
Y <- as.vector(A %*% u + sigma.e * rnorm(n.obs))We can compute kriging predictions of the process
based on these observations. To specify which locations that should be
predicted, the argument Aprd is used. This argument should
be an observation matrix that links the mesh locations to the prediction
locations.
A <- make_A(op, loc = obs.loc)
Aprd <- make_A(op, loc = proj$lattice$loc)
u.krig <- predict(op, A = A, Aprd = Aprd, Y = Y, sigma.e = sigma.e)The process simulation, and the kriging prediction are shown in the following figure.
data.df <- data.frame(x = obs.loc[, 1], y = obs.loc[, 2], field = Y, type = "Data")
krig.df <- data.frame(
x = proj$lattice$loc[, 1], y = proj$lattice$loc[, 2],
field = as.vector(u.krig$mean), type = "Prediction"
)
df_plot <- rbind(data.df, krig.df)
ggplot(df_plot) +
aes(x = x, y = y, fill = field) +
facet_wrap(~type) +
geom_raster(data = krig.df) +
geom_point(
data = data.df, aes(colour = field),
show.legend = FALSE
) +
scale_fill_viridis() +
scale_colour_viridis()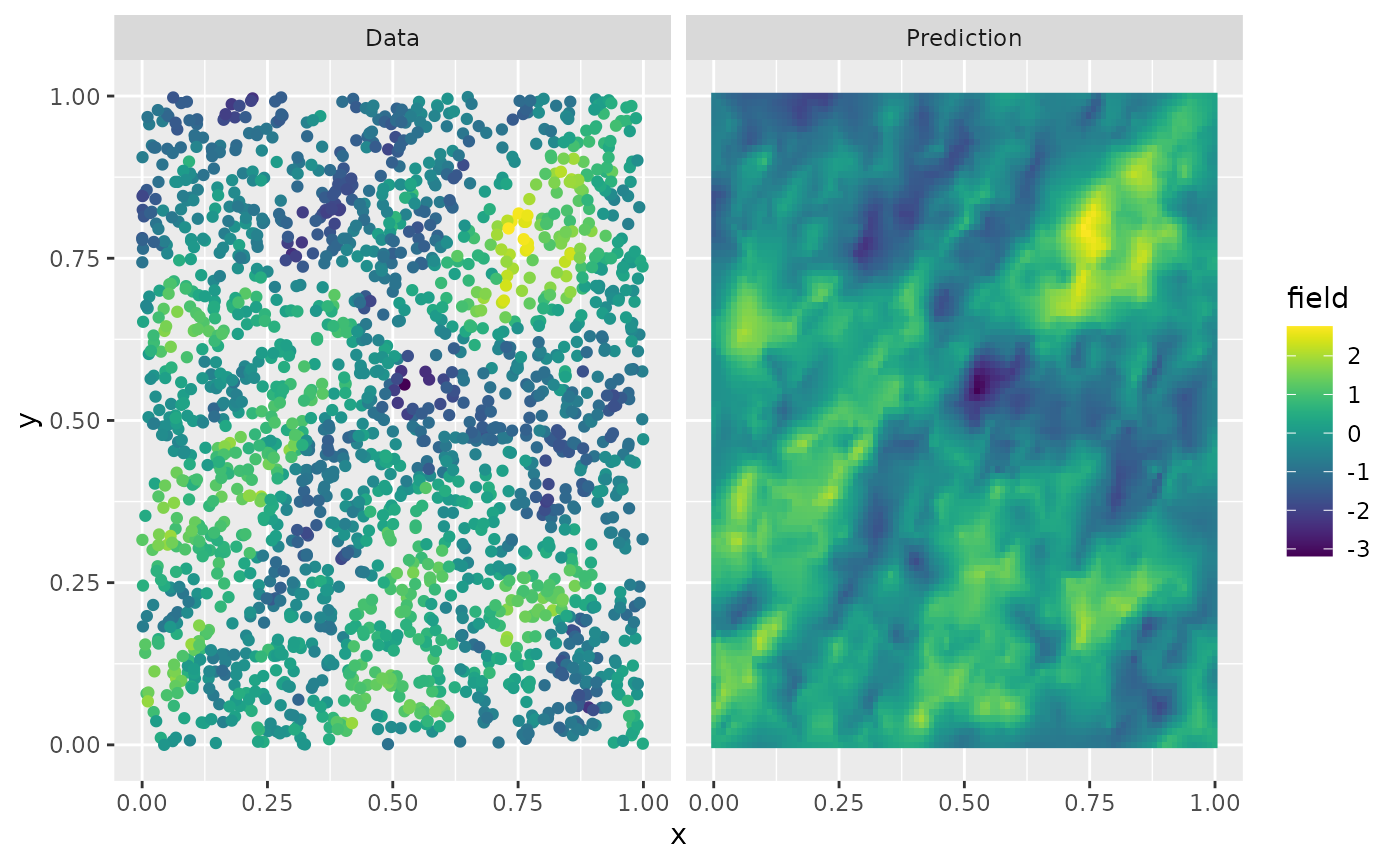 Let us finally use
Let us finally use rspde_lme to estimate the parameters
based on the data.
df <- data.frame(Y = as.matrix(Y), x = obs.loc[, 1], y = obs.loc[, 2])
res <- rspde_lme(Y ~ 1, loc = c("x", "y"), data = df, model = op, parallel = TRUE)In the call, y~1 indicates that we also want to estimate
a mean value of the model, and the arguments loc provides
the names of the spatial and temporal coordinates in the data frame. Let
us see a summary of the fitted model:
summary(res)
#>
#> Latent model - Anisotropic Whittle-Matern
#>
#> Call:
#> rspde_lme(formula = Y ~ 1, loc = c("x", "y"), data = df, model = op,
#> parallel = TRUE)
#>
#> Fixed effects:
#> Estimate Std.error z-value Pr(>|z|)
#> (Intercept) -0.1569 0.1457 -1.077 0.282
#>
#> Random effects:
#> Estimate Std.error z-value
#> nu 0.44190 0.06000 7.365
#> sigma 0.95961 0.05450 17.609
#> hx 0.08968 0.01500 5.977
#> hy 0.08914 0.01478 6.030
#> hxy 0.57242 0.04927 11.618
#>
#> Measurement error:
#> Estimate Std.error z-value
#> std. dev 0.1003 0.0027 37.15
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Log-Likelihood: -3.354359
#> Number of function calls by 'optim' = 46
#> Optimization method used in 'optim' = L-BFGS-B
#>
#> Time used to: fit the model = 26.44847 secs
#> set up the parallelization = 2.77985 secsLet us compare the estimated results with the true values:
results <- data.frame(
sigma = c(sigma, res$coeff$random_effects[2]),
hx = c(hx, res$coeff$random_effects[3]),
hy = c(hy, res$coeff$random_effects[4]),
hxy = c(hxy, res$coeff$random_effects[5]),
nu = c(nu, res$coeff$random_effects[1]),
sigma.e = c(sigma.e, res$coeff$measurement_error),
intercept = c(0, res$coeff$fixed_effects),
row.names = c("True", "Estimate")
)
print(results)
#> sigma hx hy hxy nu sigma.e
#> True 1.0000000 0.08000000 0.08000000 0.5000000 0.5000000 0.1000000
#> Estimate 0.9596148 0.08967586 0.08913957 0.5724202 0.4419019 0.1003199
#> intercept
#> True 0.0000000
#> Estimate -0.1568538Finally, we can also do prediction based on the fitted model as
pred.data <- data.frame(x = proj$lattice$loc[, 1], y = proj$lattice$loc[, 2])
pred <- predict(res, newdata = pred.data, loc = c("x", "y"))
data.df <- data.frame(x = obs.loc[, 1], y = obs.loc[, 2], field = Y, type = "Data")
krig.df <- data.frame(
x = proj$lattice$loc[, 1], y = proj$lattice$loc[, 2],
field = as.vector(u.krig$mean), type = "Prediction"
)
df_plot <- rbind(data.df, krig.df)
ggplot(df_plot) +
aes(x = x, y = y, fill = field) +
facet_wrap(~type) +
geom_raster(data = krig.df) +
geom_point(
data = data.df, aes(colour = field),
show.legend = FALSE
) +
scale_fill_viridis() +
scale_colour_viridis()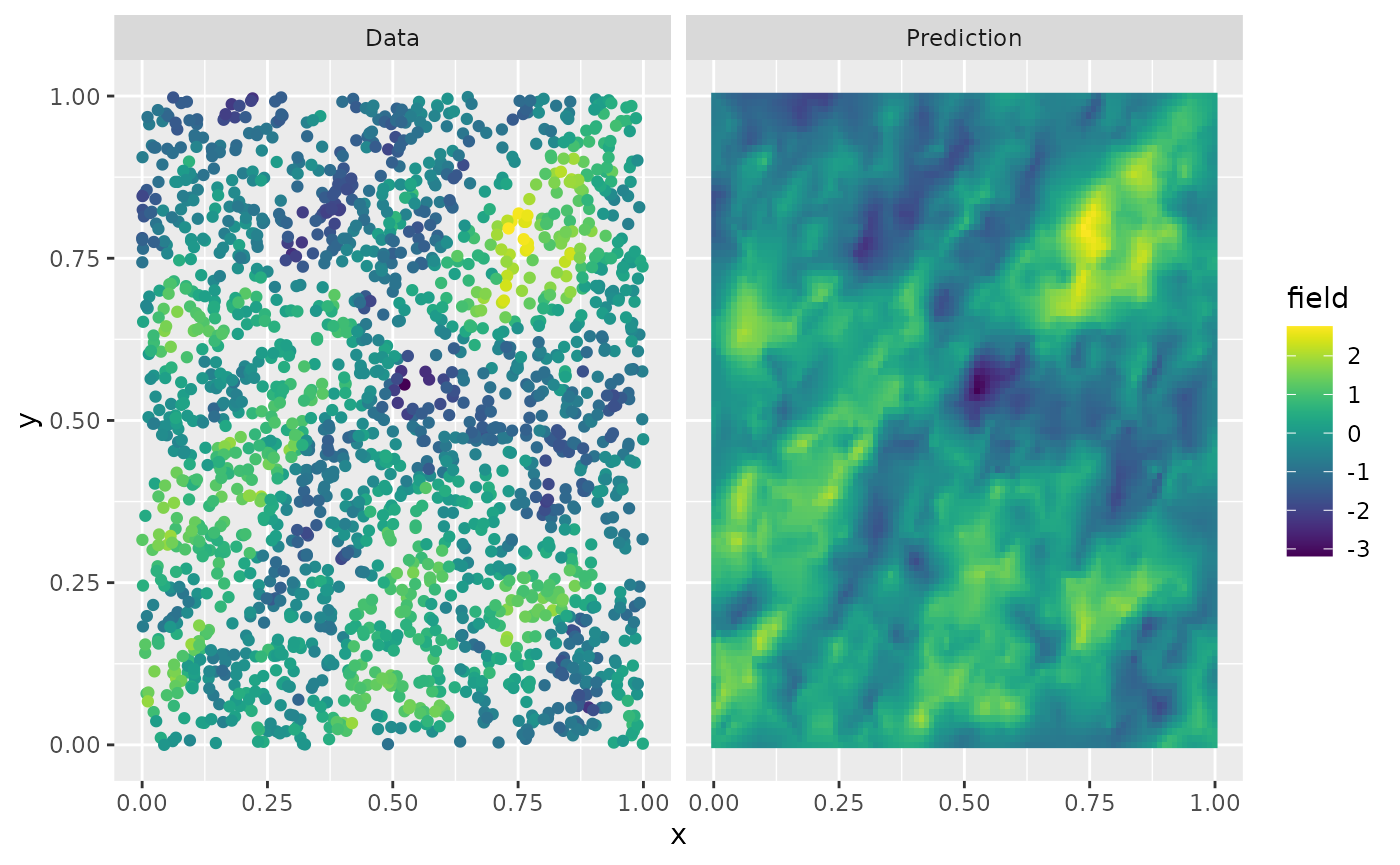
Inlabru Implementation
We will now fit the model using the inlabru package.
First, we load the package, and create the anisotropic model object:
library(inlabru)
model_aniso <- rspde.anistropic2d(mesh = mesh_2d)Then we create the component and fit the model using the
inlabru package. Observe that we can use the same data
frame as in the rspde_lme example.
cmp <- Y ~ Intercept(1) - 1 + field(cbind(x, y), model = model_aniso)
bru_fit_field <- bru(cmp, data = df, options = list(num.threads = "1:1"))Let us now compare the estimated results (the means of the
parameters) with the true values. Observe that we are using the helper
function transform_parameters_anisotropic() to change the
scale of the estimated parameters back to the original scale.
results <- data.frame(
rbind(
# True values
c(hx = hx, hy = hy, hxy = hxy, sigma = sigma, nu = nu),
# Estimated values transformed using transform_parameters_anisotropic
transform_parameters_anisotropic(
bru_fit_field$summary.hyperpar$mean[2:6],
model_aniso[["nu_upper_bound"]]
)
),
# Add sigma.e values
sigma.e = c(sigma.e, sqrt(1 / bru_fit_field$summary.hyperpar$mean[1])),
# Add intercept values
intercept = c(0, bru_fit_field$summary.fixed$mean),
row.names = c("True", "Estimate")
)
print(results)
#> hx hy hxy sigma nu sigma.e intercept
#> True 0.08 0.08 0.5 1 0.5 0.1000000 0.0000000
#> Estimate 0.09247022 0.09190848 0.5687017 0.98209 0.4559999 0.1000824 -0.1606214