
Rational approximations without finite element approximations
David Bolin and Alexandre B. Simas
Created: 2024-07-20. Last modified: 2026-01-29.
Source:vignettes/rspde_nofem.Rmd
rspde_nofem.RmdIntroduction
For models in one dimension, we can perform rational approximations without finite elements. This enables us to provide computationally efficient approximations of Gaussian processes with Mat'ern covariance functions, without having to define finite element meshes, and without having boundary effects due to the boundary conditions that are usually required. In this vignette we will introduce these methods.
We begin by loading the rSPDE package:
Assume that we want to define a model on the interval , which we want to evaluate at some locations
s <- seq(from = 0, to = 1, length.out = 101)We can now use matern.rational() to construct a rational
SPDE approximation of order
for a Gaussian random field with a Matérn covariance function on the
interval.
kappa <- 20
sigma <- 2
nu <- 0.8
r <- sqrt(8*nu)/kappa #range parameter
op_cov <- matern.rational(loc = s, nu = nu, range = r, sigma = sigma, m = 2, parameterization = "matern")The object op_cov contains the information needed for
evaluating the approximation. Note, however, that the approximation is
invariant to the locations loc, and they are only supplied
to indicate where we want to evaluate it.
To evaluate the accuracy of the approximation, let us compute the
covariance function between the process at
and all other locations in s and compare with the true
Matérn covariance function. The covariances can be calculated by using
the covariance() method.
c_cov.approx <- op_cov$covariance(ind = 1)
c.true <- matern.covariance(abs(s[1] - s), kappa, nu, sigma)The covariance function and the error compared with the Matérn covariance are shown in the following figure.
opar <- par(
mfrow = c(1, 2), mgp = c(1.3, 0.5, 0),
mar = c(2, 2, 0.5, 0.5) + 0.1
)
plot(s, c.true,
type = "l", ylab = "C(|s-0.5|)", xlab = "s", ylim = c(0, 5),
cex.main = 0.8, cex.axis = 0.8, cex.lab = 0.8
)
lines(s, c_cov.approx, col = 2)
legend("topright",
bty = "n",
legend = c("Matérn", "Rational"),
col = c("black", "red"),
lty = rep(1, 2), ncol = 1,
cex = 0.8
)
plot(s, c.true - c_cov.approx,
type = "l", ylab = "Error", xlab = "s",
cex.main = 0.8, cex.axis = 0.8, cex.lab = 0.8
)
par(opar)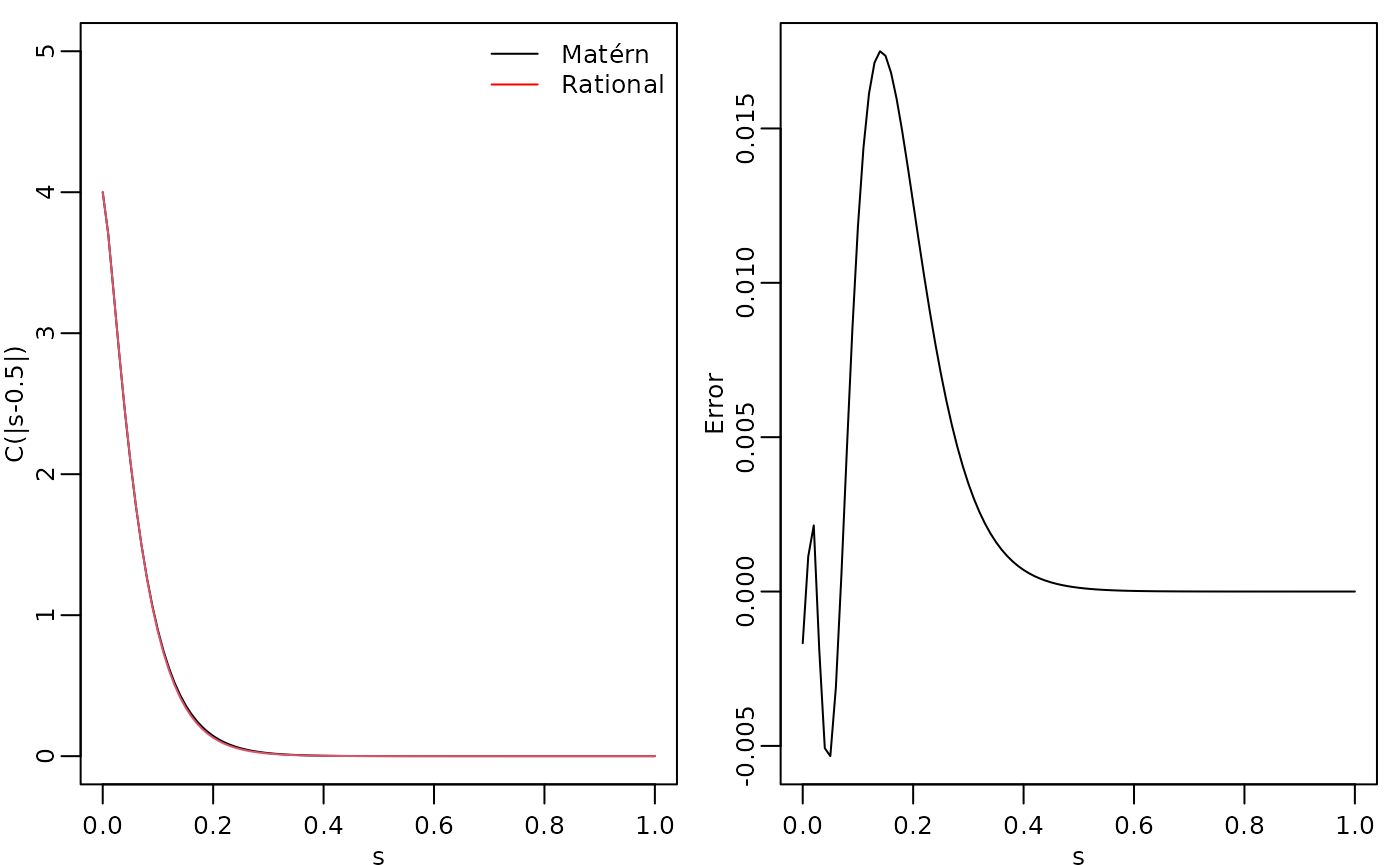
To improve the approximation we can increase the order of the approximation, by increasing . Let us, for example, compute the approximation with .
errors <- rep(0, 4)
for (i in 1:4) {
op_cov_i <- matern.rational(loc = s, range = r, sigma = sigma, nu = nu,
m = i, parameterization = "matern")
errors[i] <- norm(c.true - op_cov_i$covariance(ind = 1))
}
print(errors)## [1] 1.36208052 0.29200800 0.07907210 0.02597968We see that the error decreases very fast when we increase from to .
Simulation
We can simulate from the constructed model using the
simulate() method. To such an end we simply apply the
simulate() method to the object returned by the
matern.operators() function:
If we want replicates, we simply set the argument nsim
to the desired number of replicates. For instance, to generate two
replicates of the model, we simply do:
u.rep <- simulate(op_cov, nsim = 2)Inference
There is built-in support for computing log-likelihood functions and
performing kriging prediction in the rSPDE package. To
illustrate this, we use the simulation to create some noisy observations
of the process. For this, we first construct the observation matrix
linking the FEM basis functions to the locations where we want to
simulate. We first randomly generate some observation locations and then
construct the matrix.
We now generate the observations as , where is Gaussian measurement noise, is a covariate giving the observation location. We will assume that the latent process has a Matérn covariance with and :
n.rep <- 10
kappa <- 20
sigma <- 1.3
nu <- 0.8
r <- sqrt(8*nu)/kappa
op_cov <- matern.rational(loc = obs.loc, nu = nu, range = r, sigma = sigma, m = 2,
parameterization = "matern")
u <- matrix(simulate(op_cov, n = n.rep), ncol = n.rep)
sigma.e <- 0.3
x1 <- obs.loc
Y <- matrix(rep(2 - x1, n.rep), ncol = n.rep) + u + sigma.e * matrix(rnorm(n.obs*n.rep), ncol = n.rep)
repl <- rep(1:n.rep, each = n.obs)
df_data <- data.frame(y = as.vector(Y), loc = rep(obs.loc, n.rep),
x1 = rep(x1, n.rep), repl = repl)Let us create a new object to fit the model:
op_cov_est <- matern.rational(loc = obs.loc, m = 2)Let us now fit the model. To this end we will use the
rspde_lme() function:
fit <- rspde_lme(y~x1, model = op_cov_est,
repl = repl, data = df_data, loc = "loc",
parallel = TRUE)Here is the summary:
summary(fit)##
## Latent model - Whittle-Matern
##
## Call:
## rspde_lme(formula = y ~ x1, loc = "loc", data = df_data, model = op_cov_est,
## repl = repl, parallel = TRUE)
##
## Fixed effects:
## Estimate Std.error z-value Pr(>|z|)
## (Intercept) 2.0141 0.2957 6.812 9.6e-12 ***
## x1 -1.1959 0.4969 -2.407 0.0161 *
##
## Random effects:
## Estimate Std.error z-value
## nu 0.91412 0.10870 8.409
## sigma 1.41325 0.08784 16.089
## range 0.12563 0.01634 7.689
##
## Random effects (SPDE parameterization):
## Estimate Std.error z-value
## alpha 1.414116 0.108702 13.01
## tau 0.024819 0.001191 20.84
## kappa 21.525897 1.918353 11.22
##
## Measurement error:
## Estimate Std.error z-value
## std. dev 0.29205 0.01446 20.2
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Log-Likelihood: -876.9437
## Number of function calls by 'optim' = 42
## Optimization method used in 'optim' = L-BFGS-B
##
## Time used to: fit the model = 2.33278 mins
## set up the parallelization = 2.59163 secsLet us compare with the true values and compare the time:
print(data.frame(
sigma = c(sigma, fit$coeff$random_effects[2]),
range = c(r, fit$coeff$random_effects[3]),
nu = c(nu, fit$coeff$random_effects[1]),
row.names = c("Truth", "Estimates")
))## sigma range nu
## Truth 1.300000 0.1264911 0.8000000
## Estimates 1.413248 0.1256274 0.9141162
# Total time (time to fit plus time to set up the parallelization)
total_time <- fit$fitting_time + fit$time_par
print(total_time)## Time difference of 142.5588 secsKriging
Finally, we compute the kriging prediction of the process
at the locations in s based on these observations.
Let us create the data.frame with locations in which we
want to obtain the predictions. Observe that we also must provide the
values of the covariates.
s <- seq(from = 0, to = 1, length.out = 100)
df_pred <- data.frame(loc = s, x1 = s)We can now perform kriging with the predict() method.
For example, to predict at the locations for the first replicate:
u.krig <- predict(fit, newdata = df_pred, loc = "loc", which_repl = 1)The simulated process, the observed data, and the kriging prediction are shown in the following figure.
opar <- par(mgp = c(1.3, 0.5, 0), mar = c(2, 2, 0.5, 0.5) + 0.1)
plot(obs.loc, Y[,1],
ylab = "u(s)", xlab = "s",
ylim = c(min(c(min(u), min(Y))), max(c(max(u), max(Y)))),
cex.main = 0.8, cex.axis = 0.8, cex.lab = 0.8
)
lines(s, u.krig$mean, col = 2)
par(opar)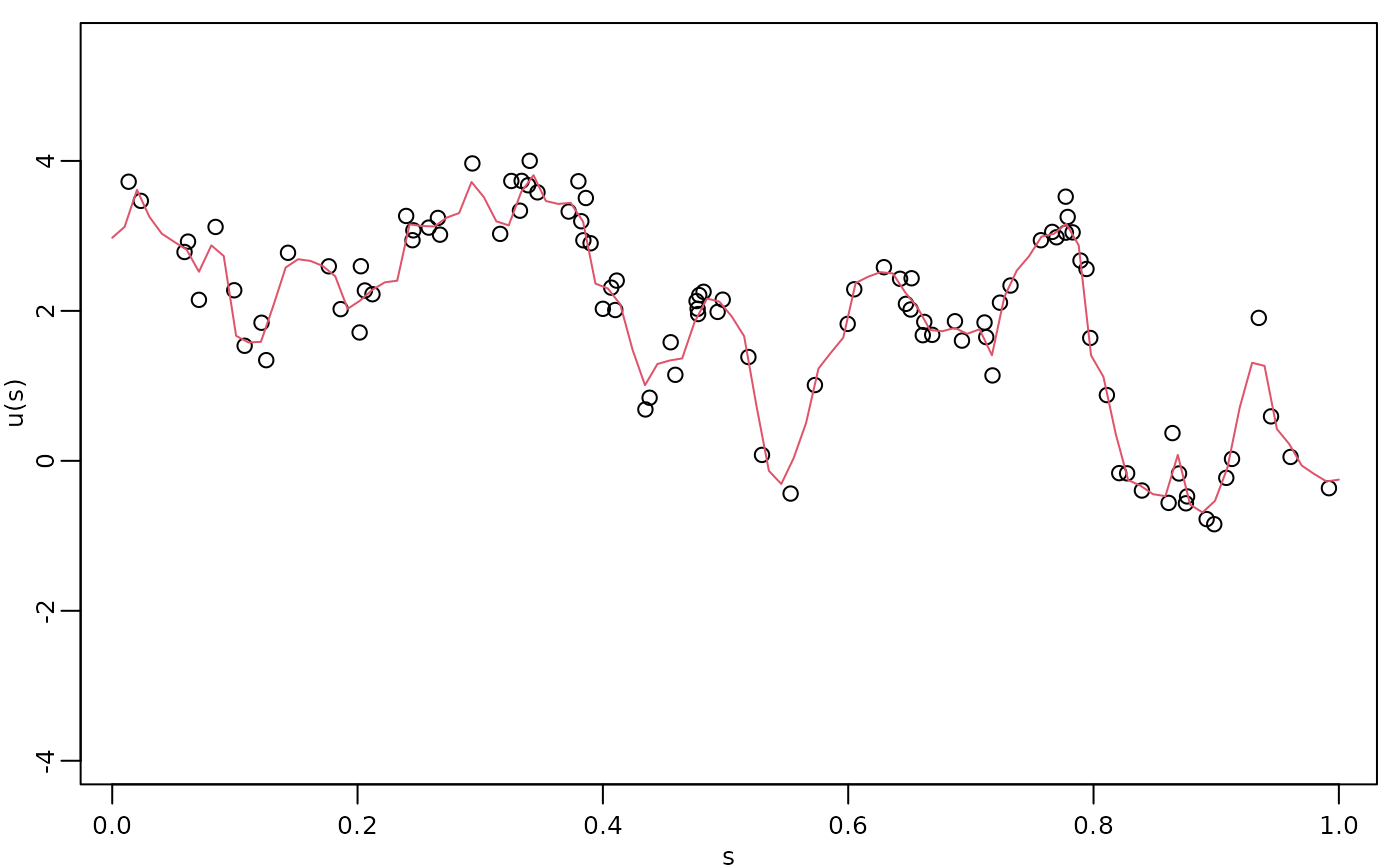
We can also use the augment() function and pipe the
results into a plot:
library(ggplot2)
library(dplyr)
augment(fit, newdata = df_pred, loc = "loc", which_repl = 1) %>% ggplot() +
aes(x = loc, y = .fitted) +
geom_line(col="red") +
geom_point(data = df_data[df_data$repl==1,], aes(x = loc, y=y))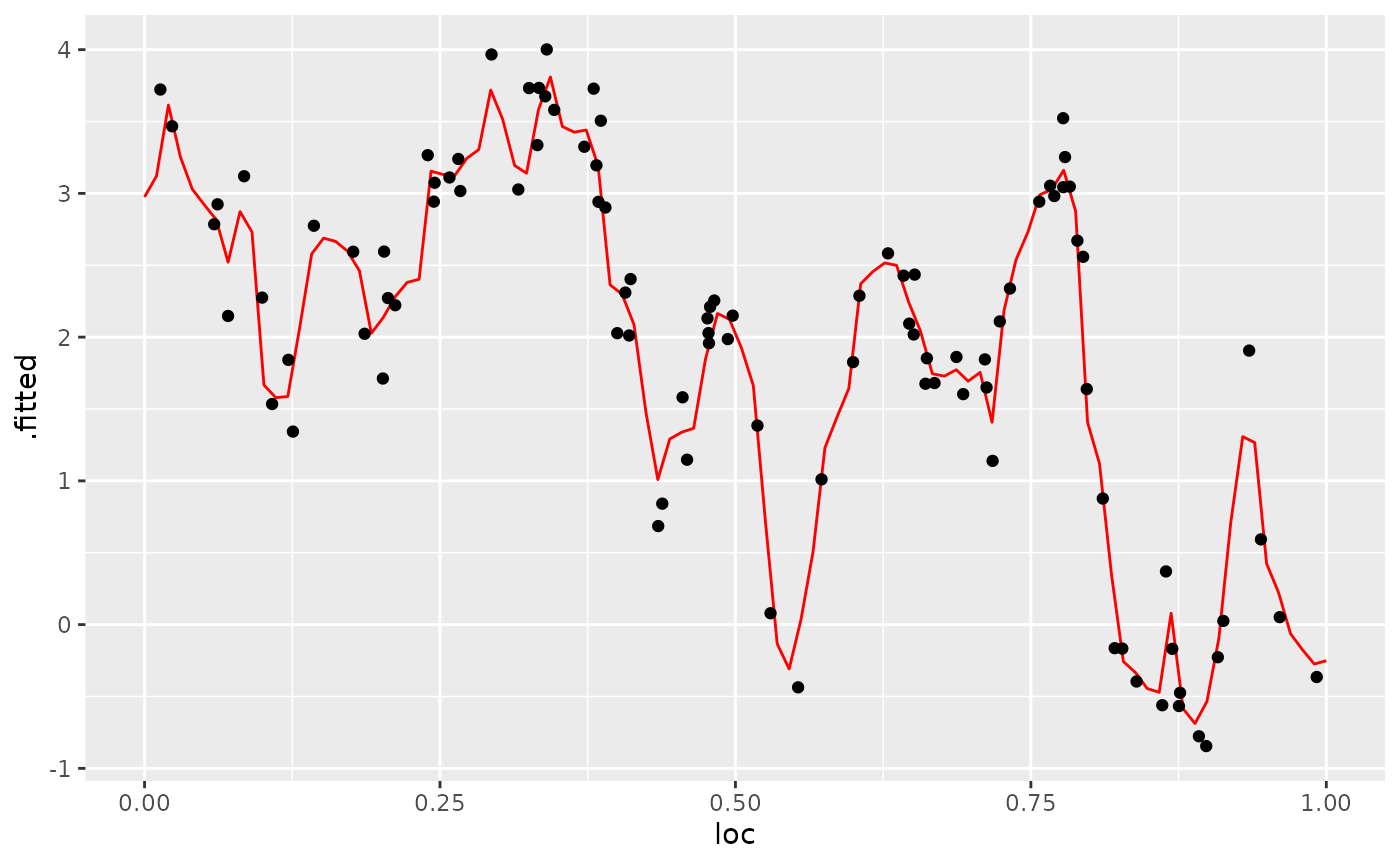
Using the models in INLA
The inla implementation of the models is found in the
rspde.matern1d function. To test it, let us simulate some
data again.
sigma <- 1
nu <- 0.8
sigma.e <- 0.1
n.obs <- 1000
r <- 1
s <- seq(from = 0, to = 5, length.out = n.obs)
op_cov <- matern.rational(loc = s, nu = nu, range = r, sigma = sigma, m = 1, parameterization = "matern")
u <- simulate(op_cov)
Y <- u + sigma.e * rnorm(n.obs)
plot(s, u, type = "l")
points(s,Y,col=2)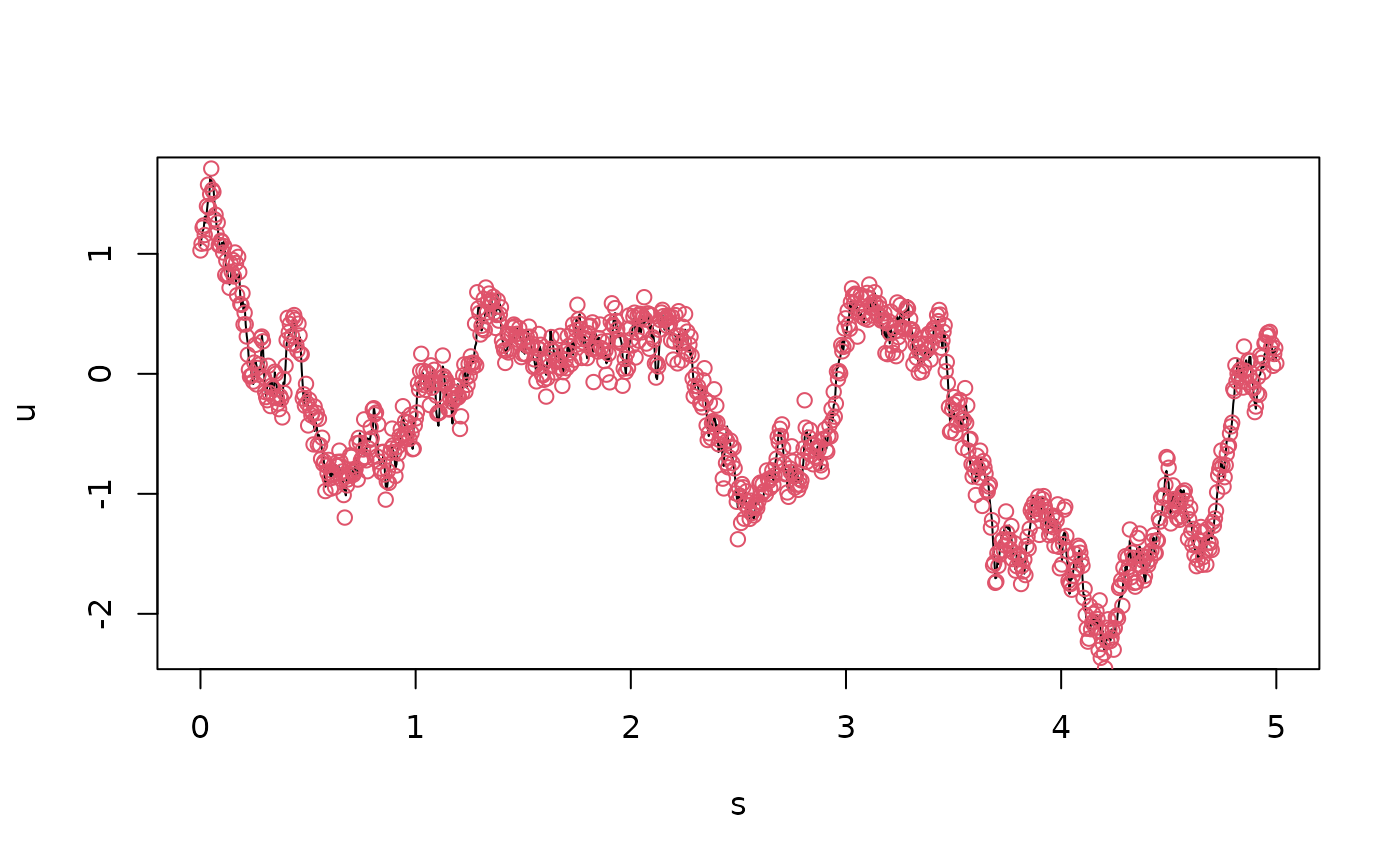 We can now specify the model and fit it to the data using
We can now specify the model and fit it to the data using
inla. In the following code, we fix the smoothness
parameter to 0.5. In this case, it will not be estimated in
inla. By removing the specification of nu in
the rspde.matern1d call, it is assumed to be free and will
be estimated from data. Note that the A matrix and the
index vector needed by inla are pre-computed
in the function. The default name of the model component (set in the
index vector) is field.
library(INLA)##
inla_model <- rspde.matern1d(loc = s, nu = 0.5)
st.dat <- inla.stack(data = list(y = as.vector(Y)), A = inla_model$A, effects = inla_model$index)
res <- inla(y ~ -1 + f(field, model = inla_model),
data = inla.stack.data(st.dat),
family = "gaussian",
control.predictor = list(A = inla.stack.A(st.dat)))Let us look at some summaries of the fit:
result_fit <- rspde.result(res, "field", inla_model, parameterization = "matern")
summary(result_fit)## mean sd 0.025quant 0.5quant 0.975quant mode
## std.dev 1.01743 0.333061 0.583143 0.943803 1.87129 0.818131
## range 2.90711 2.285620 0.778036 2.237800 9.16213 1.537500
posterior_df_fit <- gg_df(result_fit)
ggplot(posterior_df_fit) + geom_line(aes(x = x, y = y)) +
facet_wrap(~parameter, scales = "free") + labs(y = "Density")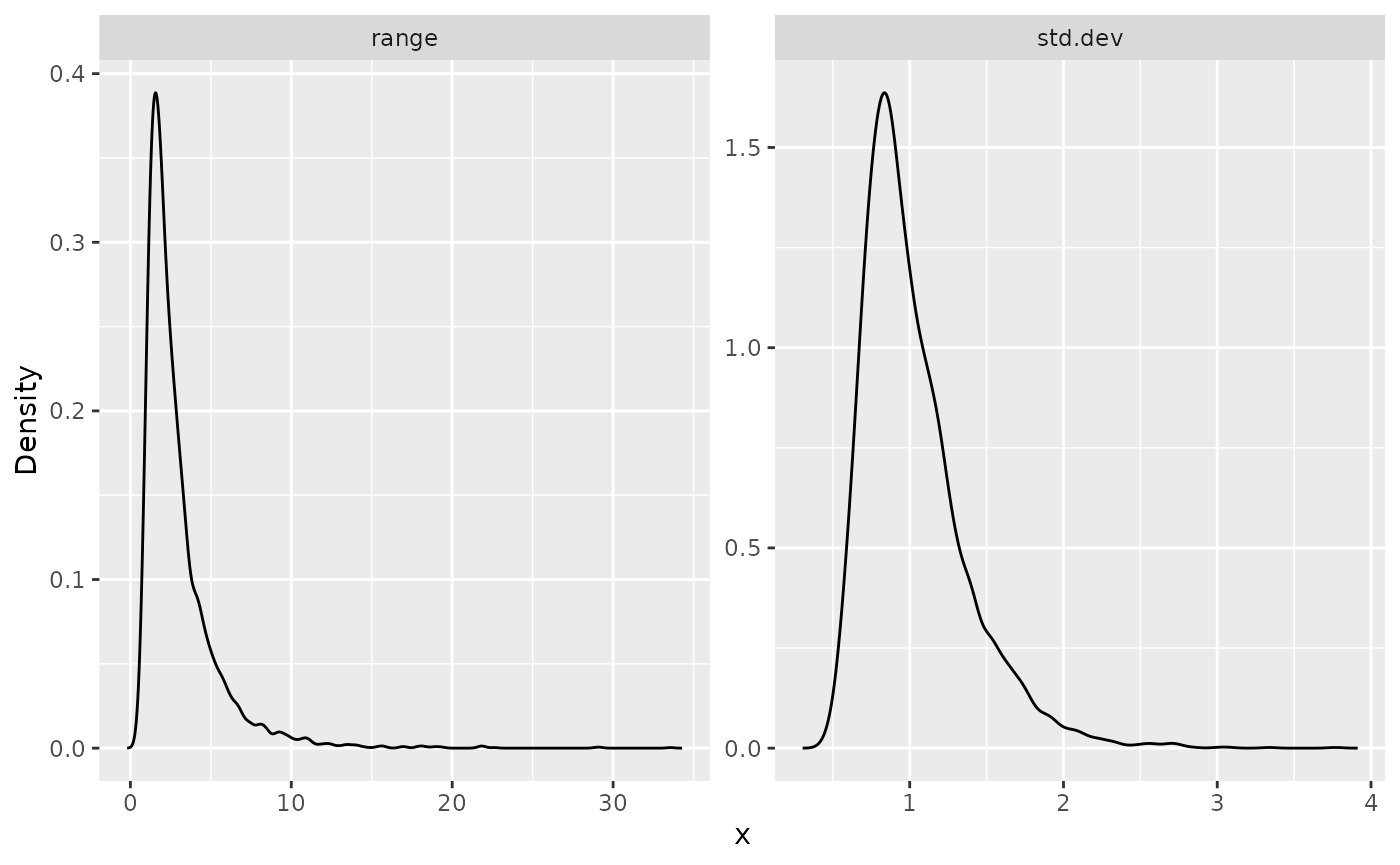
Using the models in inlabru
Let us now fit the following model using our inlabru
implementation. In this case, we will estimate nu.
sigma <- 1
nu <- 0.8
sigma.e <- 0.1
n.obs <- 251
r <- 1
s <- seq(from = 0, to = 5, length.out = n.obs)
op_cov <- matern.rational(loc = s, nu = nu, range = r, sigma = sigma, m = 1, parameterization = "matern")
u <- simulate(op_cov)
Y <- u + sigma.e * rnorm(n.obs)We need to create a data.frame with the response
variables and locations:
df_bru <- data.frame(y = Y, loc = s)We now create the model object, without specifying
nu:
bru_model <- rspde.matern1d(loc = s)Finally, we can load the inlabru package, create the
model component, and fit the model:
## Loading required package: fmesher
cmp <- y ~ -1 + Intercept(1) + field(loc, model = bru_model)
bru_fit <- bru(cmp, data = df_bru, options = list(num.threads = "1:1"))Let us, again, look at the summaries of the fit:
result_fit <- rspde.result(bru_fit, "field", bru_model, parameterization = "matern")
summary(result_fit)## mean sd 0.025quant 0.5quant 0.975quant mode
## std.dev 0.433974 0.200593 0.118527 0.411375 0.891879 0.378555
## range 0.992575 0.345106 0.466664 0.944744 1.801190 0.896720
## nu 0.865764 0.219277 0.467585 0.856026 1.313500 0.829032Kriging with the inlabru implementation
We will now do prediction with the previous model. To this end, we
start by creating the data.frame containing the locations
in which we want to do prediction:
s_pred <- seq(from = 0, to = 5, length.out = 1001)
df_pred <- data.frame(loc = s_pred)Now, first observe that we cannot directly use inlabru’s
predict method:
pred <- predict(bru_fit, newdata = df_pred, formula = ~ Intercept + field)## Error in `match_with_tolerance()`:
## ! Error: The input location 0.0050000000 is not present in the original locations used to create the model object.Indeed, it complained that we are trying to obtain predictions at
locations that were not used to create the model object. However, we can
obtain predictions using our modified predict method, in
which we need to pass the components and well as the model:
pred <- predict(bru_model, cmp, bru_fit, newdata = df_pred, formula = ~ Intercept + field)